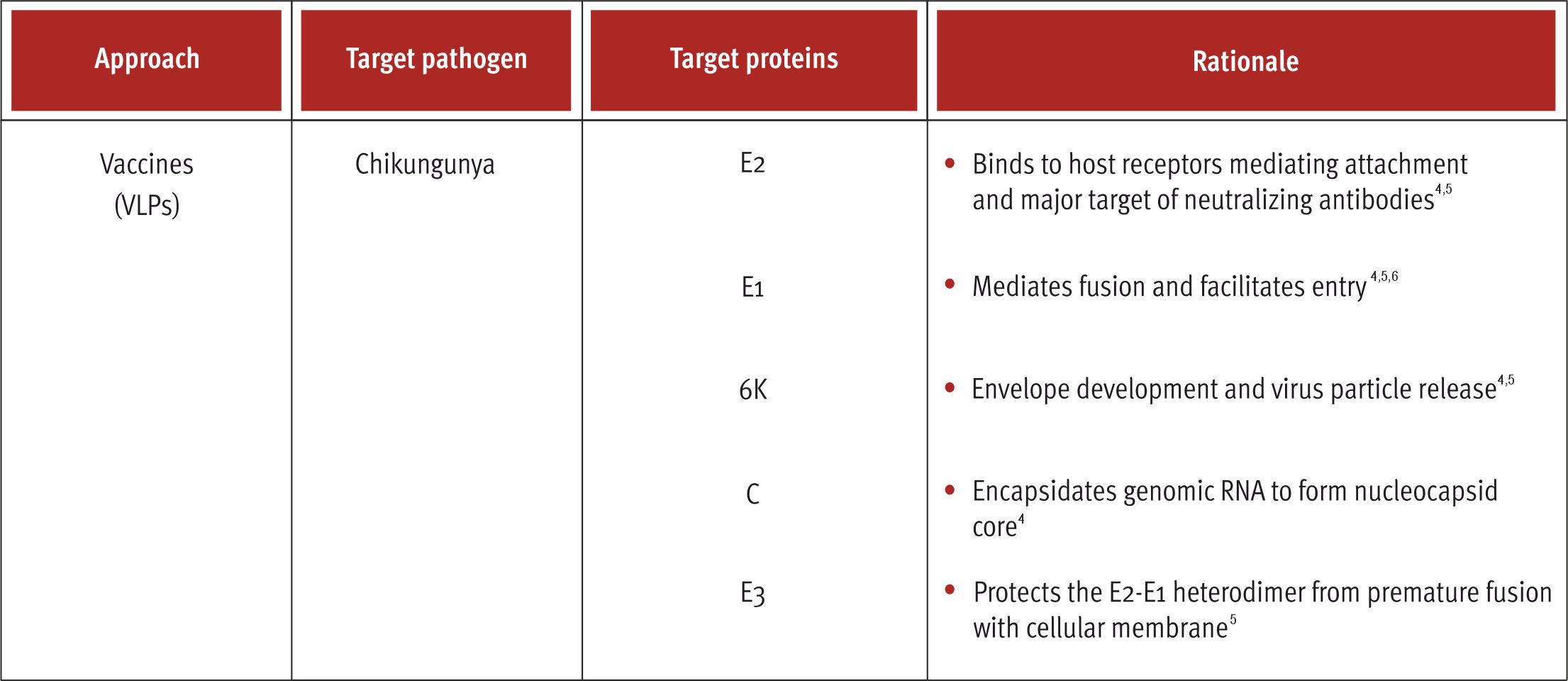
Virus-Like Particles
VLPs are multiprotein structures that mimic the organization and structure of the authentic native virus and can be produced in an array of expression systems viz. mammals, plants, bacteria, insects, and plants1,2 They are devoid of the viral genome which blunts the potential for replication within the target cells1,2. As a result, it offers improved safety and can be administered to the immunocompromised and elderly population.1,2 They are highly organized and can be self-assembled into different geometric symmetry (icosahedral, helical symmetry, rod-shape structure, and globular in shape) depending the source of the virus.3 The organized and the underlying geometry of the VLPs make them resemble pathogen-associated structural proteins (PASP) which can be efficiently recognized by the cells of the immune system.2 The size of VLPs range from 20 to 200 nm which is optimal to drain them freely in the lymphatic nodes and facilitates easy uptake the antigen-presenting cells (APCs) and presentation by the major histocompatibility complex (MHC) class II molecules.3
The potency of VLPs can elicit a protective immune response at lower antigen doses which can significantly reduce the cost of the vaccine.1
References:
- Roldão A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Virus-like particles in vaccine development. Expert Rev Vaccines. 2010;9(10):1149-1176. doi:10.1586/erv.10.115.
- Nooraei S, Bahrulolum H, Hoseini ZS, et al. Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J Nanobiotechnology. 2021;19(1):59. Published 2021 Feb 25. doi:10.1186/s12951-021-00806-7
- Tariq H, Batool S, Asif S, Ali M, Abbasi BH. Virus-Like Particles: Revolutionary Platforms for Developing Vaccines Against Emerging Infectious Diseases. Front Microbiol. 2022;12:790121. Published 2022 Jan 3. doi:10.3389/fmicb.2021.790121
- Silva LA, Dermody TS. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J Clin Invest. 2017;127(3): 737-749. doi:10.1172/JCl84417.
- Singh A, Kumar A, Yadav R, Uversky VN, Giri R. Deciphering the dark proteome of Chikungunya virus. Sci Rep. 2018;8(1):5822. Published 2018 Apr 11. doi: 10.1038/s41598-018-23969-o.
- Kuo SC, Chen YJ, WangYM, et al. Cell-based analysis ofChikungunya virus E1 protein in membrane fusion. J Biomed Sci. 2012;19(1):44. Published 2012 Apr 21. doi:10.1186/1423-0127-19-44.
